First complete genome sequences of representative strains of all Xanthomonas translucens pathovars
We assembled complete genome sequences for relevant strains of these important pathogens of cereal crops and forage grasses.
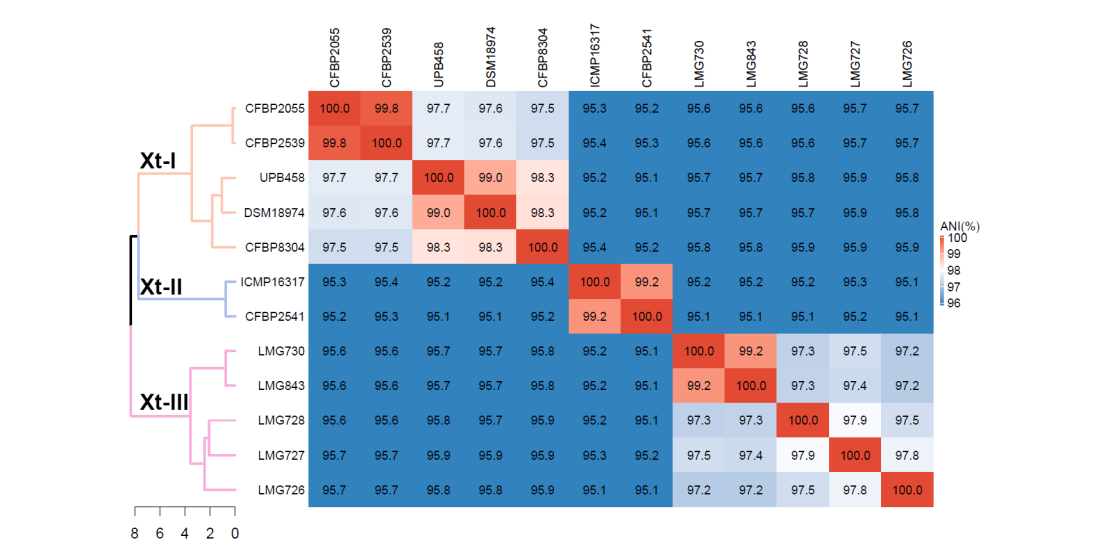
Xanthomonas translucens is a species of bacteria that can cause serious damage to cereals, such as wheat and barley, to forage grasses, as well as to pistachio. Based on the host range of the different strains, the species is divided into eleven pathovars, and two major groups are historically distinguished. The “translucens” group contains the pathovars that infect cereals and pistachio, and the “graminis” group contains the pathovars that infect forage grasses. To understand the evolutionary history of these pathovars, and to identify genes that shape their host range and are responsible for their virulence, high-quality genome sequences are a valuable resource. To date, such resources were available for only three out of the eleven pathovars of X. translucens, hindering further understanding of the host-pathogen interaction and the development of disease-resistant cultivars.
Using high-coverage long-read sequencing technology, we were able to assemble complete genome sequences for all representative strains. These allowed us to build a robust phylogeny, showing that the species is genetically divided into three clades, contrasting with the historical distinction of only two groups. Additionally, this showed some limitations of the current classification, as some pathovars showed very high similarity with each other, indicating that they could be considered as the same pathovar. Furthermore, we identified 2,181 genes that were shared by all strains and constitute the core genome of the species. In addition, we found a total of 190, 588 and 168 genes that were exclusive to each of the three clade, respectively. Investigation of type-III effectors, which are common virulence factors in Xanthomonas, allowed to identify effectors that were specific to one clade or strain, which could be involved in their pathogenicity and/or host adaptation.
Our study substantially increased the number of complete genome sequences available for X. translucens, providing high-quality genomic resources for all pathovars of the species. The virulence features of X. translucens that we identified will help to better understand the biology of each clade and/or pathovar, and processes of adaptation to their respective hosts, laying the foundations for the development of new resistant cereal crop and forage grass cultivars.
Citation:
Goettelmann F, Roman Reyna V, Cunnac S, Jacobs J M, Bragard C, Studer B, Koebnik R and Kölliker R (2022) Complete genome assemblies of all Xanthomonas translucens pathotype strains reveal three genetically distinct clades. Frontiers in Microbiology 12:817815.
external pagehttps://www.frontiersin.org/articles/10.3389/fmicb.2021.817815call_made
