ForageTILLING - Reverse Genetics in perennial ryegrass
Contact person: Chloé Manzanares
Partners: Steven Yates, Stephen Byrne, Torben Asp and Bruno Studer
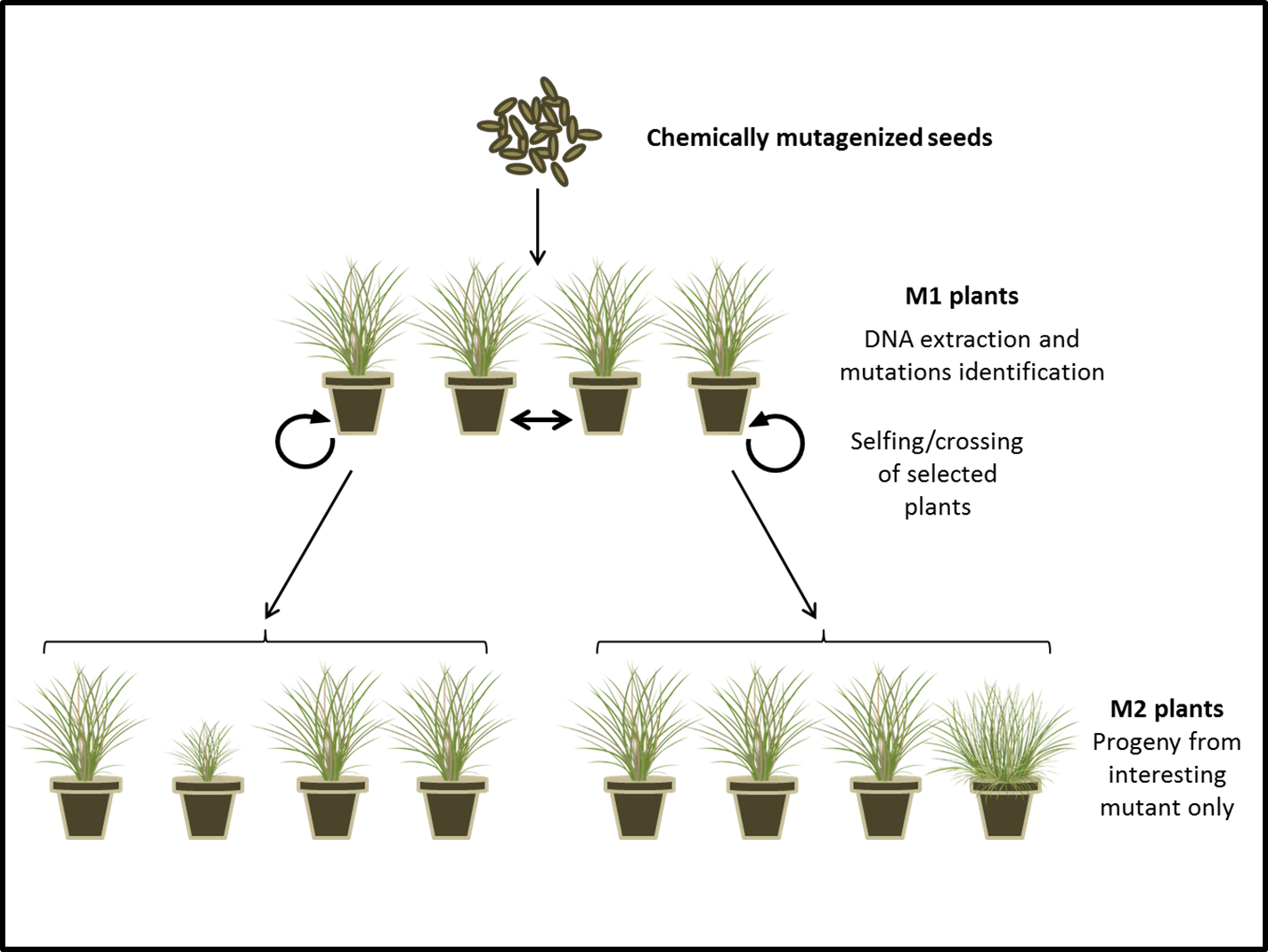
Project description: Until recently, most research projects on perennial ryegrass genetic were based on forward genetic, where a gene or genic region responsible for a phenotype was identified and mapped. However, with the genome on its way and by using synteny with other grasses, it is possible nowadays to identify gene function more precisely. Reverse genetic is a method that looks at the phenotype engendered by a specific DNA sequence. Among the reverse genetic tools, gene transformation was the most popular. However, genetic modification (GM) is not the ideal tool when working with Lolium species and breeding improvement due to the GM regulation and the difficulties linked to regeneration within these species. TILLING (Targeting Induced Local Lesions in Genomes) has evolved as a non-transgenic method to create mutants by inducing random mutations to a large number of plants, using chemical mutagen. This important functional genomic tool has been used in model species such as Arabidopsis thaliana, as well as in major crop species such as rice, wheat, maize and barley. As a reverse genetics approach, it allows the identification of an induced mutation in a specific gene of interest and to directly associate that mutation to its corresponding phenotype.
The project will focus on the construction of a perennial ryegrass TILLING population as a novel tool to identify important agronomical genes. In order to identify the mutants, the TILLING population will be screened using a targeted re-sequencing approach for genes controlling e.g. plant size, disease resistance or plant architecture. Any mutant will be self-pollinated in order to exhibit the phenotype of the homozygous mutations and characterize the gene function. Moreover, interesting phenotypes resulting from the TILLING approach can be used in breeding programs in order to improve varieties for a particular trait.