Map-based cloning of the S self-incompatibility locus
Contact person: Chloé Manzanares
Partners: Steven Yates, Daniel Thorogood, Stephen Byrne, Torben Asp, Susanne Barth and Bruno Studer
Project description: Self-incompatibility (SI) is an evolution of sexual plant reproduction, shared by more than half of the Angiosperms. SI is a system preventing pollination by “self” pollen and therefore, promoting outbreeding and genetic diversity. The mechanism used to prevent inbreeding varies between different plant groups; three main SI systems have been well characterized to day, and are genetically governed by a single S-locus. However, the SI system involved in grasses is unique as it is controlled by two loci, S and Z mapped in linkage group (LG) 1 and 2 respectively and the mechanism remains elusive to genetic and physiological levels.
Perennial ryegrass (Lolium perenne L.) is a self-incompatible tufted grass which is used in this project to map the S locus. The grass SI system is gametophytically controlled: at least one of the S or Z alleles as to be different between pollen and stigma for a compatible pollination, which can lead to different compatibility levels (see the microphotograph of pollinated stigma).
In breeding programmes, SI can be used as a tool to promote cross-pollination but it can also be a constraint, by restraining the compatible pollination and the creation of pure lines. The identification of the genetic controls as well as the physiological response cascade would enable to better control SI in breeding programme and use it at the breeder’s advantage.
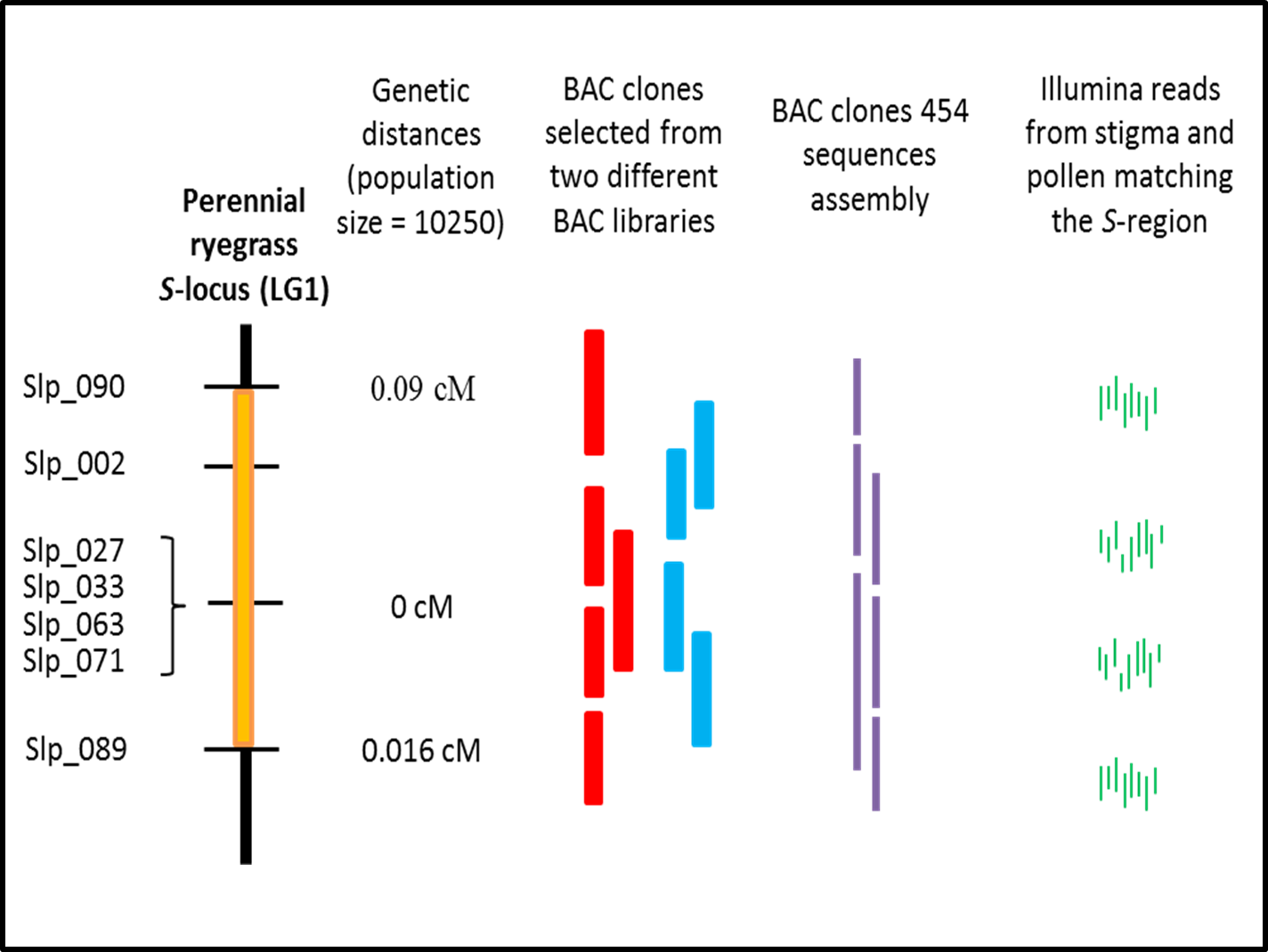
The project focuses on mapping the S-locus in perennial ryegrass and identifying the genes involved. Through a large scale fine-mapping approach combined with a fast genotyping method (High Resolution Melting (HRM) curve analysis), the S-locus region was reduced down to 0.11cM on LG1. This genomic region was sequenced and annotated in order to assess the genes within it and transcripts from pollen and stigma were sequenced in order to study the gene expression in these regions as well as the allelic diversity. The S-locus project identified four stigma S-candidate genes and potentially six pollen S-candidate genes.